6. Miniconda¶
Conda is an open source package management system and environment management system. Conda allows you find and install packages in your own environment. You don’t need administrator privileges.
If you need a package that requires a different version of Python, you use conda as an environment manager. With just a few commands, you can set up a totally separate environment to run that different version of Python, while continuing to run your usual version of Python in your normal environment.
6.1. Installation¶
basic install is 89M
other packages will require additional space, so rather install it in
/zfs/omics/personal/<uvanetid1>than in your/home/<uvanetid1>
6.1.1. Get miniconda¶
Download it
cd /zfs/omics/personal/${USER}
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
6.1.2. Install Miniconda¶
Run the install script (as you)
bash Miniconda3-latest-Linux-x86_64.sh
Read the license information and answer yes….. After you answered yes, you will see a message like:
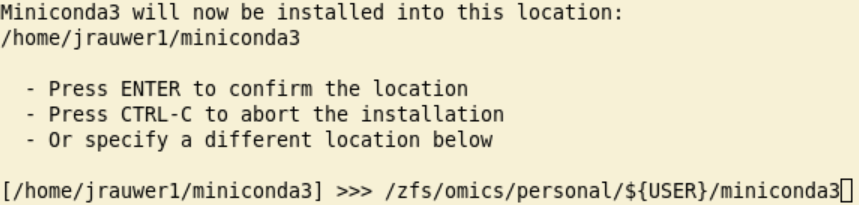
Specify here
/zfs/omics/personal/${USER}/miniconda3(or replace\${USER}with your uvanetid).When the installation has finished you are asked if you want to initialize conda. Answer yes:
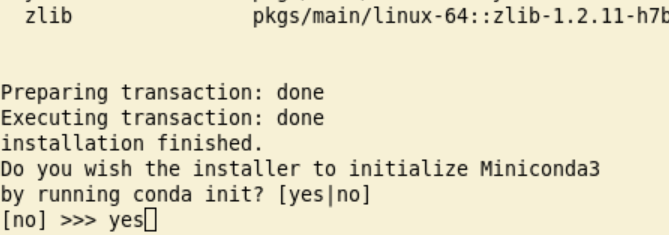
This finishes the installation:
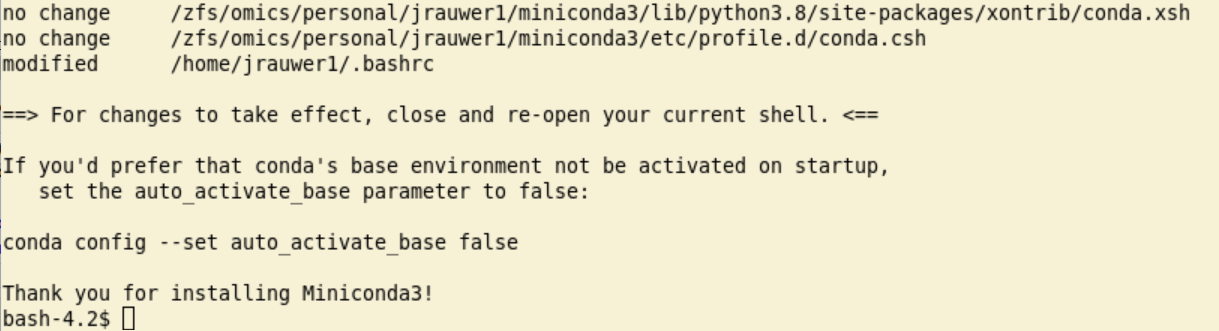
After the installation, to have a shell with conda, leave your current one and start a new one or type:
source ~/.bashrc
After you have done this, you will see (base) as a prefix in your shell prompt.
So, each time you start a shell, conda will be activated, if you don’t want that, type:
conda config --set auto_activate_base fals
How much space does miniconda3 take?
cd /zfs/personal/${USER}/
du -hs miniconda3/
#437M miniconda3/
6.2. Installation of a Conda Package¶
Suppose, you want to make a bacterial assembly with flye. With
which flyeyou discover that flye is not installed on Crunchomics. Of course, you can ask the maintainers of Crunchomics to install flye. But you also could install flye as a conda package. You go to https://anaconda.org/ and search for flyeThere is a conda package available:
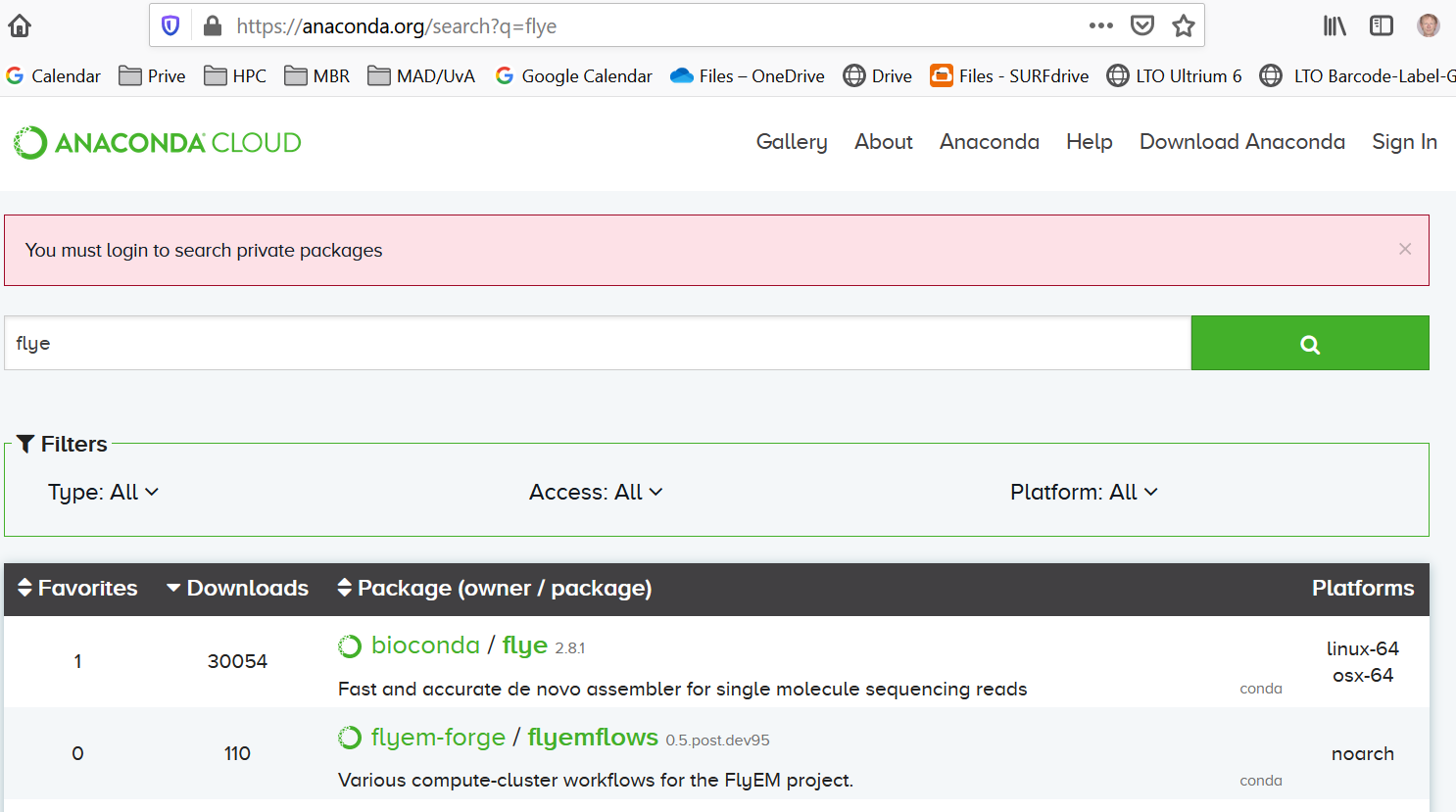
When you click on flye you get a link with installation details:
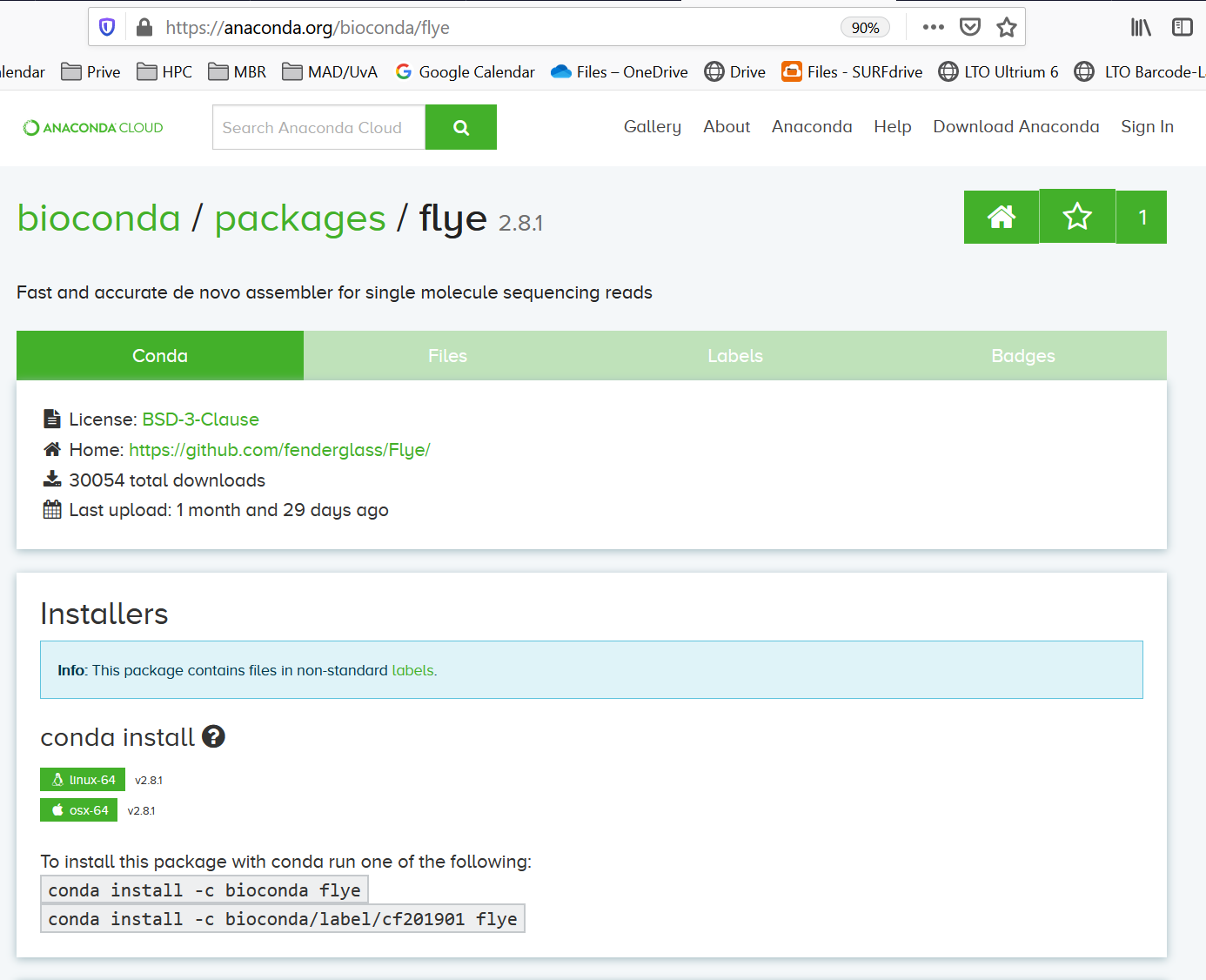
Suppose you don’t want flye in your base environment. Therefore, you create a new environment that you call nptools. Install flye in the nptools evironment, deactivate base and activate nptools:
which flye #indeed, no flye
conda create -n nptools
conda install -n nptools -c bioconda flye
conda deactivate
conda activate nptools
flye -h
#usage: flye (--pacbio-raw | --pacbio-corr | --pacbio-hifi | --nano-raw |
# --nano-corr | --subassemblies) file1 [file_2 ...]
# --genome-size SIZE --out-dir PATH
conda env list
conda deactivate
flye
#bash: flye: command not found
#
du -hs /zfs/personal/${USER}/miniconda3/
#820M miniconda3/
Look what is in your environment with:
conda list -n nptools
conda list -n base
6.2.1. Example: Using Conda Installation of Flye¶
conda deactivate
conda activate nptools
cd /zfs/omics/personal/${USER}/
mkdir -p ecoli
cd ecoli
wget https://zenodo.org/record/1172816/files/Loman_E.coli_MAP006-1_2D_50x.fasta
date
flye --nano-raw Loman_E.coli_MAP006-1_2D_50x.fasta --out-dir . --threads 4
date